rad-plot-tb2j-magnons.py#
Script for plotting magnon dispersion from TB2J "exchange.out" file.
New in version 0.7.12.
Script plots magnon dispersion spectra following the algorithm described in Magnon dispersion.
It requires TB2J output file "exchange.out" and the information about the magnetic ground state.
Example is based on the files from examples folder.
Ground state input#
Ground state is defined by two sets of parameters:
- Directions of spin in the unit cell (or supercell)
To define direction of spins in the supercell use -s, --spin parameter. You need to use the names of the atoms in the "exchange.out" file. Each name has to be followed by three numbers, separated by spaces. The numbers represent the x, y, and z components of the spin vector:
-s Cr1 0 0 1.5 Cr2 0 0 1.5
- Spin-spiral vector. (single-Q incommensurate structure).
Spin spiral is defined by two parameters, each parameter is a vector:
- \(\boldsymbol{\vec{Q}}\) (-Q, --spiral-vector)
It is relative to the reciprocal cell.
- Global rotation axis \(\boldsymbol{\vec{n}}\) (-ra, --rotation-axis)
It is given in absolute coordinate in a real space. Only the direction of the vector matters.
In the input file "exchange.out" six atoms are present: Br1, Cr1, S1, Br2, Cr2, S2. Only Cr1 and Cr2 have exchange interaction between them. Therefore, it is necessary to specify spin vectors for Cr1 and Cr2. You can specify spin vectors for all atoms, but it is not necessary.
Custom k-points#
By default you can use predefined k-points based on the high symmetry points for each Bravais lattice. However, you can specify your own high symmetry points using -kps, --k-points argument:
rad-plot-tb2j-magnons.py -if exchange.out -kps X1 "$\mathrm{X_1}$" -0.5 0 0 Y1 "$\mathrm{Y_1}$" 0 -0.5 0 -kp X1-G-Y1 -s Cr1 0 0 1.5 Cr2 0 0 1.5 -on CrSBr-custom-kpoints
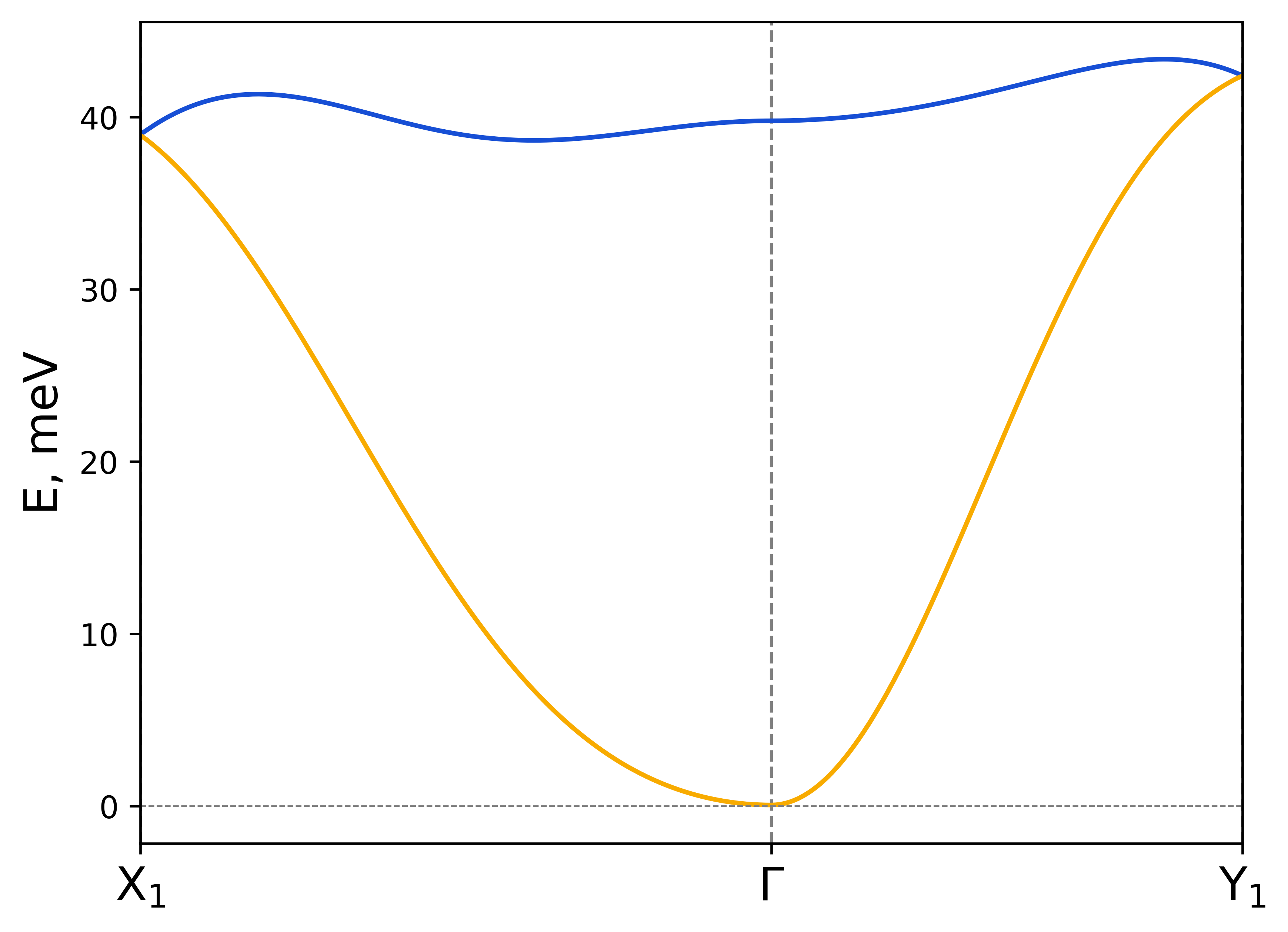
The syntax is the following:
-kps <name1> <label1> <kx1> <ky1> <kz1> <name2> <label2> <kx2> <ky2> <kz2> ...
where:
- <name> is the name of the k-point
It can be used in the -kp, --k-path argument.
Note
If an error like:
ValueError: Point 'X' already defined.
appears, then you need to use different name for the k-points, because the name "X" is already used. Only the name is checked for uniqueness. Therefore, you can provide the same label for different k-points or custom label for one of the predefined k-points.
- <label> is the label of the k-point
This string is directly passed to the plot. You can use LaTeX syntax here.
<kx> <ky> <kz> are the coordinates of the k-point relative to the reciprocal cell.
Template file#
Exchange template file (see Template specification) can be used to form the model or to filter the spin Hamiltonian. Formation of the model means that the exchange parameters from the TB2J are averaged over the individual bonds following the specification of the template.
Filtering of the model#
For filtering the spin Hamiltonian there are a few options available:
Control of parameters#
You can "turn off" parts of the full exchange matrix:
- -noa/--no-anisotropic
Ignore anisotropic symmetric exchange in the spinham.
Examples#
CrSBr#
CrSBr is a ferromagnet with two magnetic sites in the unit cell. The minimum input is the "exchange.out" file and the spin vectors for the two Cr atoms:
rad-plot-tb2j-magnons.py -if exchange.out -s Cr1 0 0 1.5 Cr2 0 0 1.5 -on CrSBr
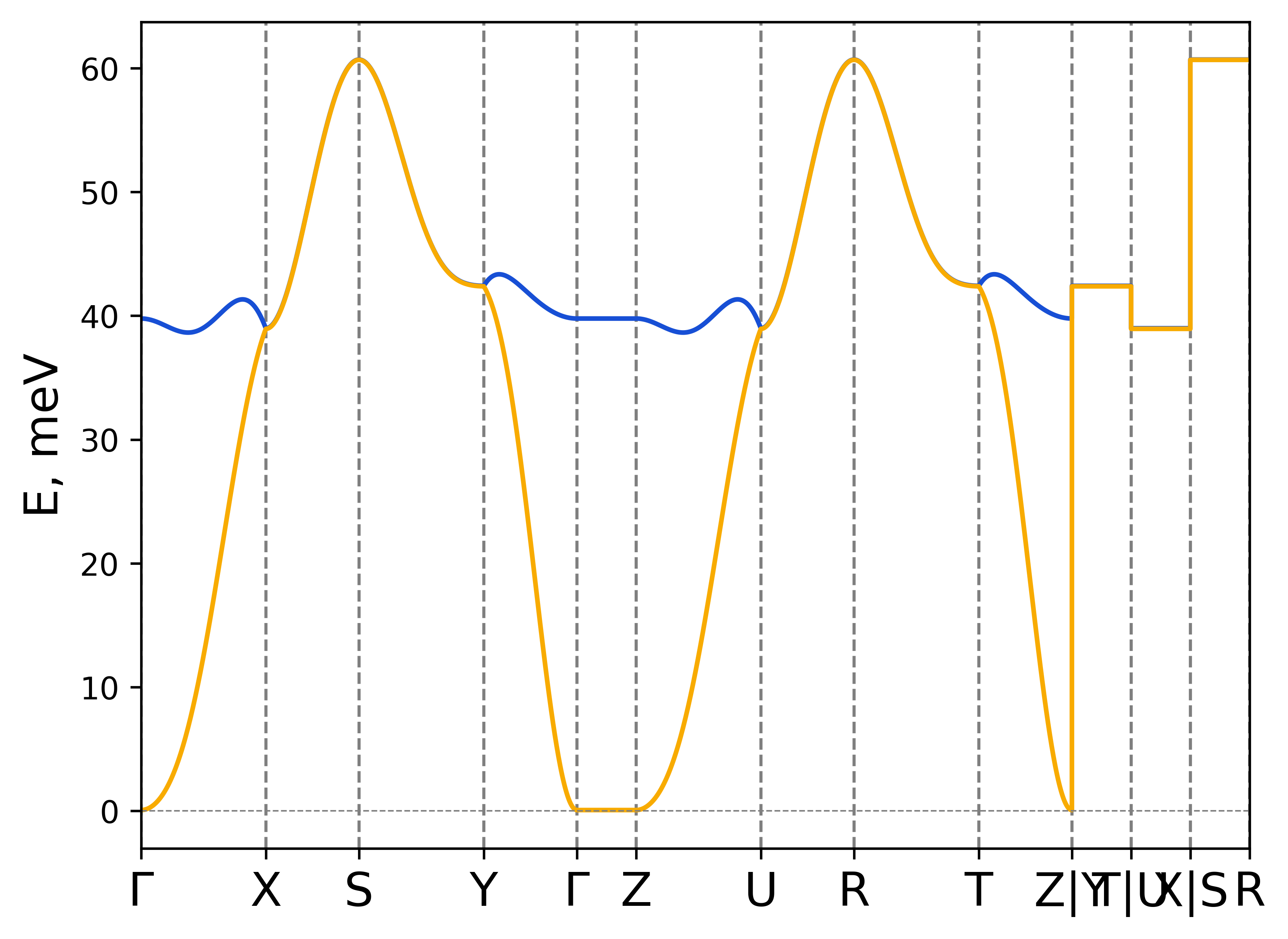
Magnon dispersion for CrSBr.#
Default k-path was detected based on the symmetry (Orthorhombic (ORC)) of the lattice.
Hint
Sometimes due to numerical inaccuracies detected symmetry of the lattice is not the one that you expect. In that case you may want to use --bravais-type argument. With this argument the script tries to reduce numerical accuracy to match the desired Bravais lattice type. If it fails, then check your lattice, maybe it is not the one you expect.
You can customise the k-path using --k-path argument (see Path for details):
rad-plot-tb2j-magnons.py -if exchange.out -s Cr1 0 0 1.5 Cr2 0 0 1.5 -kp G-X-S-Y-G -on CrSBr-custom-path
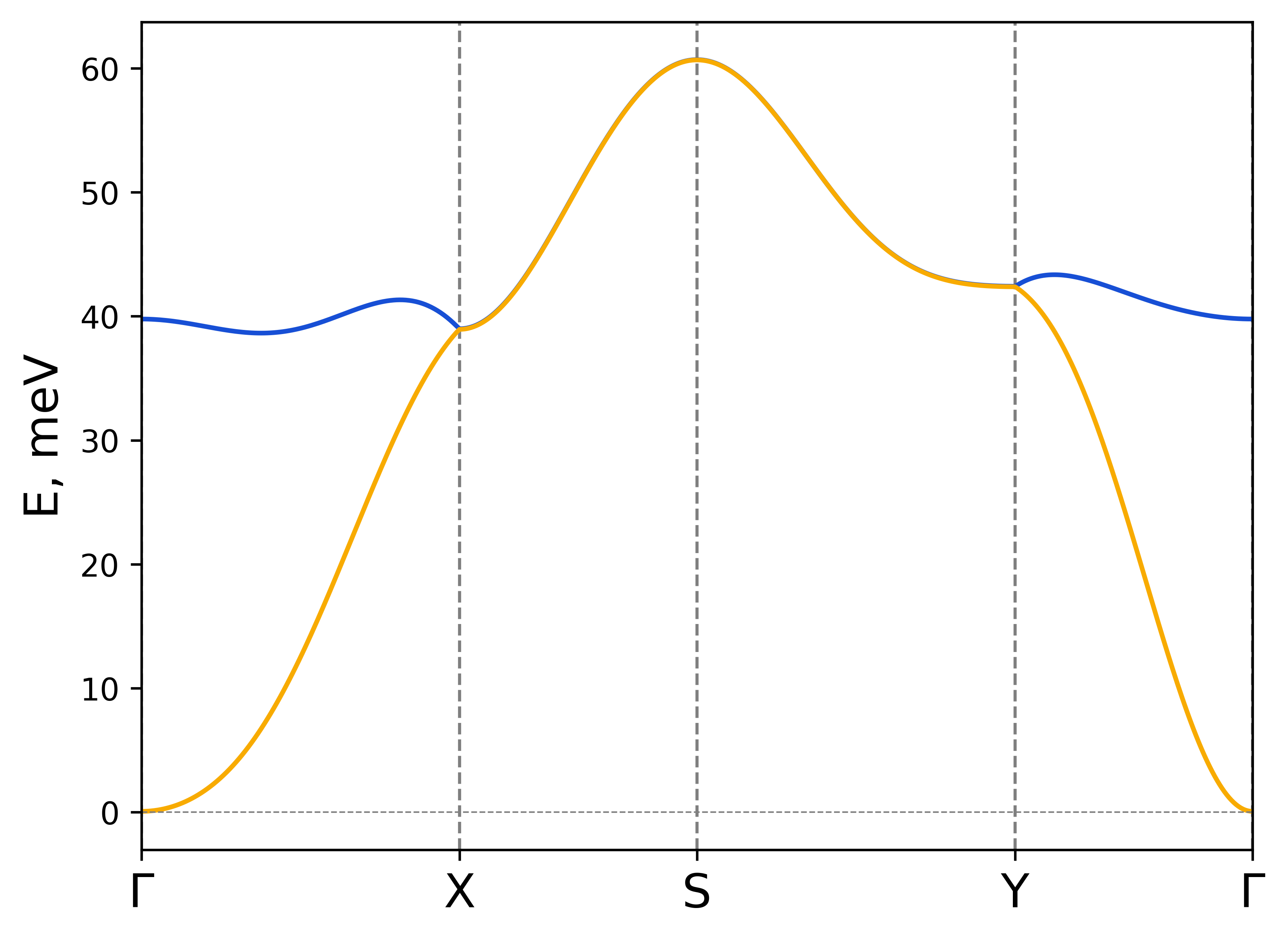
Magnon dispersion for CrSBr with custom path.#
Arguments#
-if, --input-filename#
Relative or absolute path to the "exchange.out" file, including the name and extension of the file itself.
required
type: str
-s, --spin#
Spin of the atoms in the model.
For each atom, which has at least one bond connected to it is necessary to specify spin vector. The spin vector is specified in the form of atom's name followed by three numbers, separated by spaces. The numbers represent the x, y, and z components of the spin vector.
required
type: list of str
-tf, --template-file#
Relative or absolute path to the template file, including the name and extension of the file.
optional
type: str
-on, --output-name#
Seedname for the output files.
default: "magnon_dispersion"
type: str
-Q, --spiral-vector#
Spin spiral vector. Relative to the reciprocal cell.
optional
type: list of 3 float
-ra, --rotation-axis#
Direction of global rotation axis. In absolute coordinates in real space.
optional
type: list of 3 float
-kps, --k-points#
Additional high-symmetry k-points.
Coordinates are relative to the reciprocal cell.
optional
type: list of str
New in version 0.8.9.
-kp, --k-path#
Path in reciprocal space for the magnon dispersion.
optional
type: str
-fm, --form-model#
Whether to form the spinham based on the template.
default: False
type: bool
-R, --R-vector#
R vectors for filtering the spin Hamiltonian.
In TB2J outputs the bond is defined by atom 1 (from) and atom 2 (to). Atom 1 is always located in (0, 0, 0) unit cell, while atom 2 is located in R = (i, j, k) unit cell. This parameter tells the script to keep only the bonds for which atom 2 is located in one of specified R supercells. Supercells are specified by a set of integers separated by spaces. They are grouped by three starting from the left and forms a set of R vectors. If the last group contains 1 or 2 integers they are ignored.
optional
type: list of int
-maxd, --max-distance#
(<=) Maximum distance.
All the bonds with the distance between atom 1 and atom 2 greater than maximum distance are excluded from the model.
optional
type: float
New in version 0.8.0.
-mind, --min-distance#
(>=) Minimum distance.
All the bonds with the distance between atom 1 and atom 2 lower than minimum distance are excluded from the Hamiltonian.
optional
type: float
New in version 0.8.0.
-st, --save-txt#
Whether to save data to .txt file.
Two files appears: "output-name.txt" and "output-name_info.txt". First one contains raw data of the graph, second one contains information about the parameters.
default: False
type: bool
-i, --interactive#
Whether to show interactive plot.
default: False
type: bool
-v, --verbose#
Verbose output, propagates to the called methods.
default: False
type: bool
-bt, --bravais-type#
Bravais lattice type. If not provided, the type is identified automatically.
It does not force the Bravais lattice type on the model,
but tries to reach the desired type by reducing the
numerical accuracy in the lepage() algorithm.
optional
type: str
-jo, --join-output#
Whether to join the output files into a single file.
default: False
type: bool
-nodmi#
Whether to ignore DMI in the spinham.
default: False
type: bool
-noa, --no-anisotropic#
Whether to ignore anisotropic symmetric exchange in the spinham.
default: False
type: bool
